Remote sensing of plant functional traits
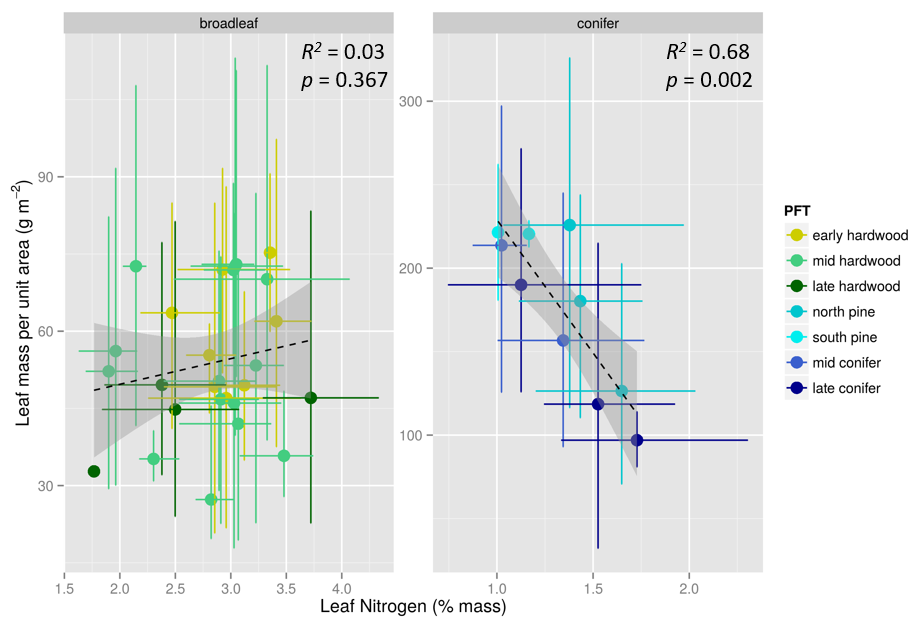
Most ecosystem models currently represent plants as belonging to discrete “functional types” with fixed parameters, but approaches based on continuously varying plant traits more effectively capture gradual adaptation of ecosystems to environmental change. Recent syntheses of large trait databases have contributed immensely to our understanding of drivers of global plant trait variability at evolutionary time scales. However, major axes of interspecific trait variability, such as the trade-off between leaf nitrogen and mass per unit area (“leaf economics spectrum”), are insufficient to represent variability in other critical aspects of plant function such as water transport. Furthermore, global trait covariance patterns do not necessarily hold at smaller scales such as within plant functional types or biomes, yet these are the scales of the most policy-relevant ecological change. Tracking spatial and temporal variability in plant traits through succession and in response to environmental change is a task uniquely suited for remote sensing due to the required spatial scale and temporal continuity of observations.
Bayesian inversion of radiative transfer models
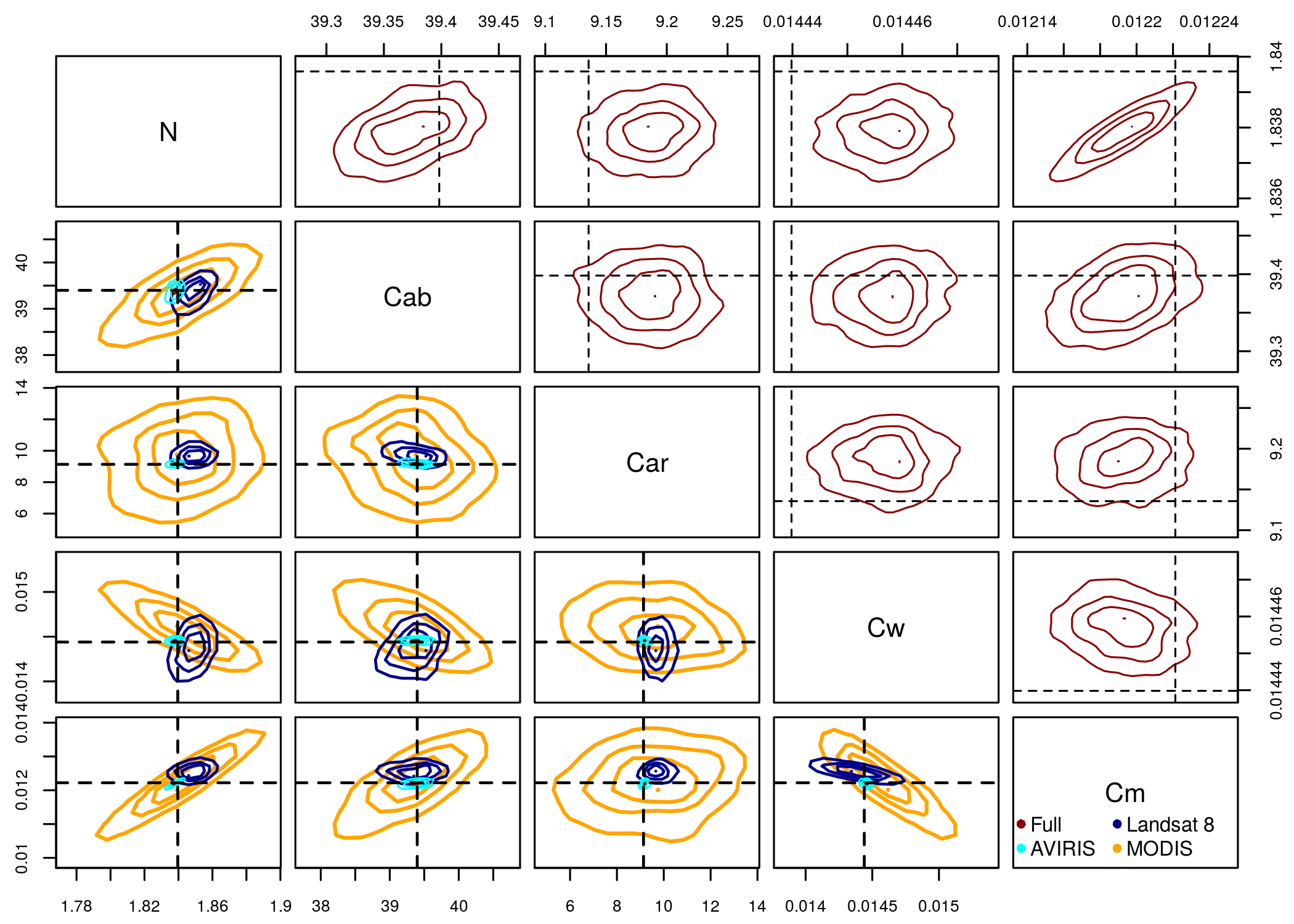
Typical approaches to remote sensing of surface characteristics involve extrapolation of empirical relationships between remotely sensed observations (e.g. surface reflectance, vegetation indices) and ground observations of the variables of interest (e.g. chlorophyll concentration, leaf area index).
However, a more general and robust approach is to use physically based radiative transfer models (RTMs), which provide a mechanistic link between surface optical properties and ecological variabiles and are therefore inherently more generalizable. My research involves applications of Bayesian statistical algorithms to retrieve RTM parameters and their uncertainties from remotely sensed reflectance observations.
PEcAn Project
PEcAn is a scientific workflow and ecoinformatics toolbox designed to facilitate the task of ecosystem modeling. Collaborators on the PEcAn project can be found at Brookhaven National Lab, University of Wisconsin, and the National Center for Supercomputing Applications (NCSA). The list of models PEcAn can handle is constantly growing. For more information, go to the project website.
My tools
The majority of our work is accomplished in R. For computationally intensive tasks, we use lower-level languages like Fortran and C++. For Bayesian inference, we use JAGS (Just Another Gibbs Sampler). Our entire lab and most of our colleagues use GitHub for version control and collaboration.
My personal toolbox mostly involves the command line, including a reasonably complicated Vim setup (eventually, I will have a post dedicated to my setup).
About this website
On this site, I will try to share tips and tricks I’ve accumulatd related to programming, data analysis, research, and graduate student life in general.
This site is operated through the Jekyll blogging system and is hosted by GitHub. My current theme of choice is a tweaked version of the zetsu theme. All of the code for this website is publicly available in my GitHub repository.